Note
Go to the end to download the full example code
Multiclass MEG ERP Decoding¶
Decoding applied to MEG data in sensor space decomposed using Xdawn. After spatial filtering, covariances matrices are estimated and classified by the MDM algorithm (Nearest centroid).
4 Xdawn spatial patterns (1 for each class) are displayed, as per the for mean-covariance matrices used by the classification algorithm.
# Authors: Alexandre Barachant <alexandre.barachant@gmail.com>
#
# License: BSD (3-clause)
import numpy as np
from matplotlib import pyplot as plt
import mne
from mne import io
from mne.datasets import sample
from sklearn.metrics import (
classification_report,
confusion_matrix,
ConfusionMatrixDisplay,
)
from sklearn.model_selection import KFold
from sklearn.pipeline import make_pipeline
from pyriemann.classification import MDM
from pyriemann.estimation import XdawnCovariances
print(__doc__)
Set parameters and read data
data_path = str(sample.data_path())
raw_fname = data_path + "/MEG/sample/sample_audvis_filt-0-40_raw.fif"
event_fname = data_path + "/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif"
tmin, tmax = -0.0, 1
event_id = dict(aud_l=1, aud_r=2, vis_l=3, vis_r=4)
# Setup for reading the raw data
raw = io.Raw(raw_fname, preload=True)
raw.filter(2, None, method="iir") # replace baselining with high-pass
events = mne.read_events(event_fname)
raw.info["bads"] = ["MEG 2443"] # set bad channels
picks = mne.pick_types(
raw.info, meg="grad", eeg=False, stim=False, eog=False, exclude="bads"
)
# Read epochs
epochs = mne.Epochs(
raw,
events,
event_id,
tmin,
tmax,
proj=False,
picks=picks,
baseline=None,
preload=True,
verbose=False,
)
labels = epochs.events[:, -1]
evoked = epochs.average()
Opening raw data file /home/docs/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Reading 0 ... 41699 = 0.000 ... 277.709 secs...
Filtering raw data in 1 contiguous segment
Setting up high-pass filter at 2 Hz
IIR filter parameters
---------------------
Butterworth highpass zero-phase (two-pass forward and reverse) non-causal filter:
- Filter order 8 (effective, after forward-backward)
- Cutoff at 2.00 Hz: -6.02 dB
Removing projector <Projection | PCA-v1, active : False, n_channels : 102>
Removing projector <Projection | PCA-v2, active : False, n_channels : 102>
Removing projector <Projection | PCA-v3, active : False, n_channels : 102>
Removing projector <Projection | Average EEG reference, active : False, n_channels : 60>
Decoding with Xdawn + MDM
n_components = 3 # pick some components
# Define a monte-carlo cross-validation generator (reduce variance):
cv = KFold(n_splits=10, shuffle=True, random_state=42)
pr = np.zeros(len(labels))
epochs_data = epochs.get_data()
print("Multiclass classification with XDAWN + MDM")
clf = make_pipeline(XdawnCovariances(n_components), MDM())
for train_idx, test_idx in cv.split(epochs_data):
y_train, y_test = labels[train_idx], labels[test_idx]
clf.fit(epochs_data[train_idx], y_train)
pr[test_idx] = clf.predict(epochs_data[test_idx])
print(classification_report(labels, pr))
/home/docs/checkouts/readthedocs.org/user_builds/pyriemann/checkouts/latest/examples/ERP/plot_classify_MEG_mdm.py:80: FutureWarning: The current default of copy=False will change to copy=True in 1.7. Set the value of copy explicitly to avoid this warning
epochs_data = epochs.get_data()
Multiclass classification with XDAWN + MDM
precision recall f1-score support
1 0.99 0.96 0.97 72
2 0.91 0.99 0.95 73
3 0.97 0.99 0.98 73
4 1.00 0.93 0.96 70
accuracy 0.97 288
macro avg 0.97 0.96 0.97 288
weighted avg 0.97 0.97 0.97 288
plot the spatial patterns
xd = XdawnCovariances(n_components)
xd.fit(epochs_data, labels)
info = evoked.copy().resample(1).info # make it 1Hz for plotting
patterns = mne.EvokedArray(
data=xd.Xd_.patterns_.T, info=info
)
patterns.plot_topomap(
times=[0, n_components, 2 * n_components, 3 * n_components],
ch_type="grad",
colorbar=False,
size=1.5,
time_format="Pattern %d"
)
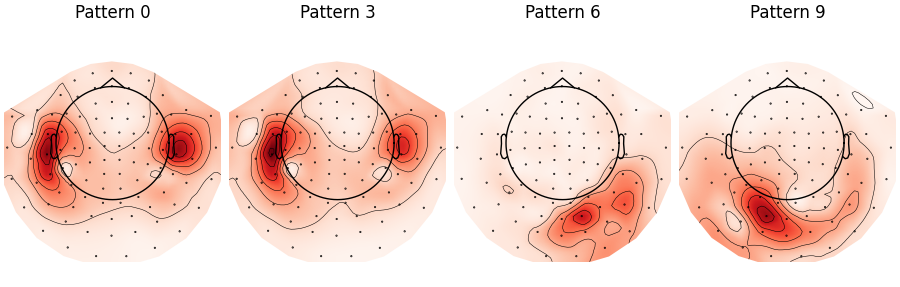
<MNEFigure size 900x287.5 with 4 Axes>
plot the confusion matrix
names = ["audio left", "audio right", "vis left", "vis right"]
cm = confusion_matrix(labels, pr)
ConfusionMatrixDisplay(cm, display_labels=names).plot()
plt.show()
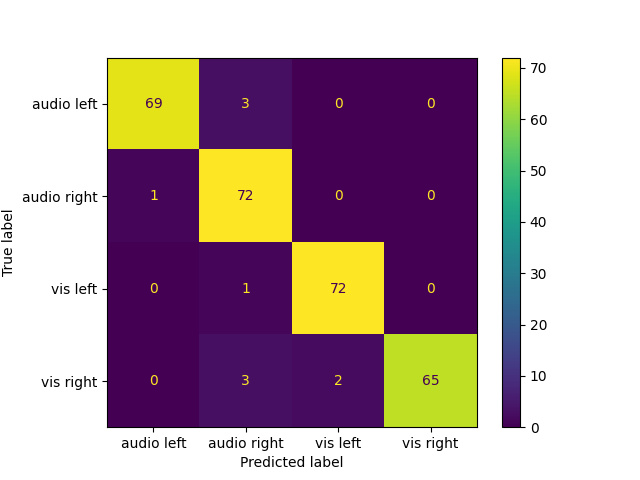
Total running time of the script: (0 minutes 11.405 seconds)