Note
Go to the end to download the full example code
Embedding ERP MEG data in 2D Euclidean space¶
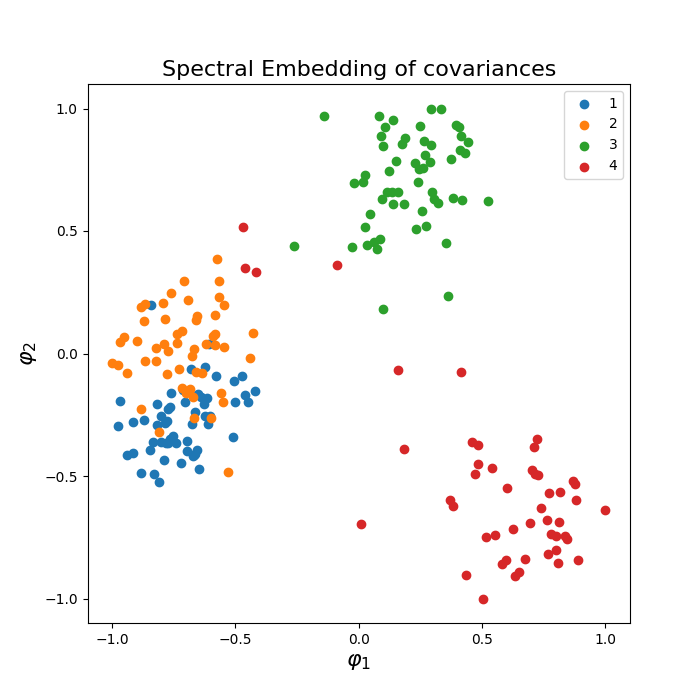
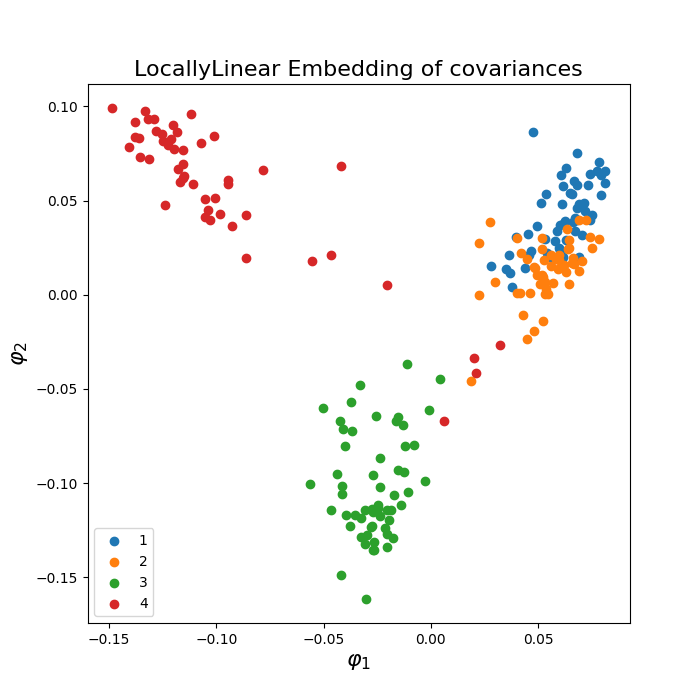
Riemannian embeddings via Laplacian Eigenmaps (LE) and Locally Linear Embedding (LLE) of a set of ERP data. Embedding via Laplacian Eigenmaps is referred to as Spectral Embedding (SE).
Locally Linear Embedding (LLE) assumes that the local neighborhood of a point on the manifold can be well approximated by the affine subspace spanned by the k-nearest neighbors of the point and finds a low-dimensional embedding of the data based on these affine approximations.
Laplacian Eigenmaps (LE) are based on computing the low dimensional representation that best preserves locality instead of local linearity in LLE [1].
# Authors: Pedro Rodrigues <pedro.rodrigues01@gmail.com>,
# Gabriel Wagner vom Berg <gabriel@bccn-berlin.de>
# License: BSD (3-clause)
import matplotlib.pyplot as plt
import mne
from mne import io
from mne.datasets import sample
from sklearn.model_selection import train_test_split
from pyriemann.estimation import XdawnCovariances
from pyriemann.utils.viz import plot_embedding
print(__doc__)
Set parameters and read data¶
data_path = str(sample.data_path())
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
event_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
tmin, tmax = -0., 1
event_id = dict(aud_l=1, aud_r=2, vis_l=3, vis_r=4)
# Setup for reading the raw data
raw = io.Raw(raw_fname, preload=True, verbose=False)
raw.filter(2, None, method='iir') # replace baselining with high-pass
events = mne.read_events(event_fname)
raw.info['bads'] = ['MEG 2443'] # set bad channels
picks = mne.pick_types(raw.info, meg=True, eeg=False, stim=False, eog=False,
exclude='bads')
# Read epochs
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, proj=False,
picks=picks, baseline=None, preload=True, verbose=False)
X = epochs.get_data(copy=False)
y = epochs.events[:, -1]
Filtering raw data in 1 contiguous segment
Setting up high-pass filter at 2 Hz
IIR filter parameters
---------------------
Butterworth highpass zero-phase (two-pass forward and reverse) non-causal filter:
- Filter order 8 (effective, after forward-backward)
- Cutoff at 2.00 Hz: -6.02 dB
Embedding of Xdawn covariance matrices¶
nfilter = 4
xdwn = XdawnCovariances(estimator='scm', nfilter=nfilter)
split = train_test_split(X, y, train_size=0.25, random_state=42)
Xtrain, Xtest, ytrain, ytest = split
covs = xdwn.fit(Xtrain, ytrain).transform(Xtest)
Laplacian Eigenmaps (LE), also called Spectral Embedding (SE)¶
plot_embedding(covs, ytest, metric='riemann', embd_type='Spectral',
normalize=True)
plt.show()
Locally Linear Embedding (LLE)¶
plot_embedding(covs, ytest, metric='riemann', embd_type='LocallyLinear',
normalize=False)
plt.show()
References¶
Total running time of the script: (0 minutes 12.343 seconds)