Note
Go to the end to download the full example code
One Way manova time¶
One way manova to compare Left vs Right in time.
from time import time
import numpy as np
from pylab import plt
import seaborn as sns
from mne import Epochs, pick_types, events_from_annotations
from mne.io import concatenate_raws
from mne.io.edf import read_raw_edf
from mne.datasets import eegbci
from pyriemann.stats import PermutationDistance
from pyriemann.estimation import Covariances
sns.set_style('whitegrid')
Set parameters and read data¶
# avoid classification of evoked responses by using epochs that start 1s after
# cue onset.
tmin, tmax = -2., 6.
event_id = dict(hands=2, feet=3)
subject = 1
runs = [6, 10, 14] # motor imagery: hands vs feet
raw_files = [
read_raw_edf(f, preload=True, verbose=False)
for f in eegbci.load_data(subject, runs)
]
raw = concatenate_raws(raw_files)
events, _ = events_from_annotations(raw, event_id=dict(T1=2, T2=3))
picks = pick_types(
raw.info, meg=False, eeg=True, stim=False, eog=False, exclude='bads')
raw.filter(7., 35., method='iir', picks=picks)
epochs = Epochs(
raw,
events,
event_id,
tmin,
tmax,
proj=True,
picks=picks,
baseline=None,
preload=True,
verbose=False)
labels = epochs.events[:, -1] - 2
# get epochs
epochs_data = epochs.get_data(copy=False)
Used Annotations descriptions: ['T1', 'T2']
Filtering raw data in 3 contiguous segments
Setting up band-pass filter from 7 - 35 Hz
IIR filter parameters
---------------------
Butterworth bandpass zero-phase (two-pass forward and reverse) non-causal filter:
- Filter order 16 (effective, after forward-backward)
- Cutoffs at 7.00, 35.00 Hz: -6.02, -6.02 dB
Pairwise distance based permutation test¶
covest = Covariances()
Fs = 160
window = 2 * Fs
Nwindow = 20
Ns = epochs_data.shape[2]
step = int((Ns - window) / Nwindow)
time_bins = range(0, Ns - window, step)
pv = []
Fv = []
# For each frequency bin, estimate the stats
t_init = time()
for t in time_bins:
covmats = covest.fit_transform(epochs_data[:, ::1, t:(t + window)])
p_test = PermutationDistance(1000, metric='riemann', mode='pairwise')
p, F = p_test.test(covmats, labels, verbose=False)
pv.append(p)
Fv.append(F[0])
duration = time() - t_init
# plot result
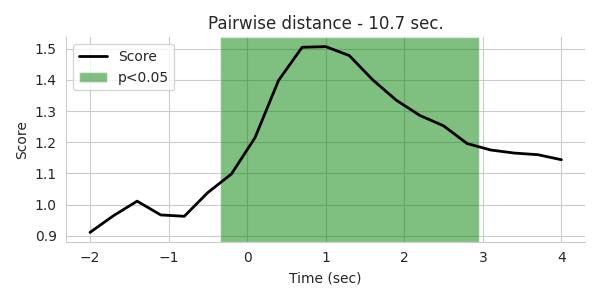
fig, axes = plt.subplots(1, 1, figsize=[6, 3], sharey=True)
sig = 0.05
times = np.array(time_bins) / float(Fs) + tmin
axes.plot(times, Fv, lw=2, c='k')
plt.xlabel('Time (sec)')
plt.ylabel('Score')
a = np.where(np.diff(np.array(pv) < sig))[0]
a = a.reshape(int(len(a) / 2), 2)
st = (times[1] - times[0]) / 2.0
for p in a:
axes.axvspan(times[p[0]] - st, times[p[1]] + st, facecolor='g', alpha=0.5)
axes.legend(['Score', 'p<%.2f' % sig])
axes.set_title('Pairwise distance - %.1f sec.' % duration)
sns.despine()
plt.tight_layout()
plt.show()
Total running time of the script: (0 minutes 11.704 seconds)