Note
Go to the end to download the full example code
Artifact Correction by AJDC-based Blind Source Separation¶
Blind source separation (BSS) based on approximate joint diagonalization of Fourier cospectra (AJDC), applied to artifact correction of EEG [1].
# Authors: Quentin Barthélemy & David Ojeda.
# EEG signal kindly shared by Marco Congedo.
#
# License: BSD (3-clause)
import gzip
import numpy as np
from matplotlib import pyplot as plt
from mne import create_info
from mne.io import RawArray
from mne.viz import plot_topomap
from mne.preprocessing import ICA
from scipy.signal import welch
from pyriemann.spatialfilters import AJDC
from pyriemann.utils.viz import plot_cospectra
def read_header(fname):
"""Read the header of sample-blinks.txt"""
with gzip.open(fname, 'rt') as f:
content = f.readline().split()
return content[:-1], int(content[-1])
Load EEG data¶
fname = '../data/sample-blinks.txt.gz'
signal_raw = np.loadtxt(fname, skiprows=1).T
ch_names, sfreq = read_header(fname)
ch_count = len(ch_names)
duration = signal_raw.shape[1] / sfreq
Channel space¶
# Plot signal X
ch_info = create_info(ch_names=ch_names, ch_types=['eeg'] * ch_count,
sfreq=sfreq)
ch_info.set_montage('standard_1020')
signal = RawArray(signal_raw, ch_info, verbose=False)
signal.plot(duration=duration, start=0, n_channels=ch_count,
scalings={'eeg': 3e1}, color={'eeg': 'steelblue'},
title='Original EEG signal', show_scalebars=False)
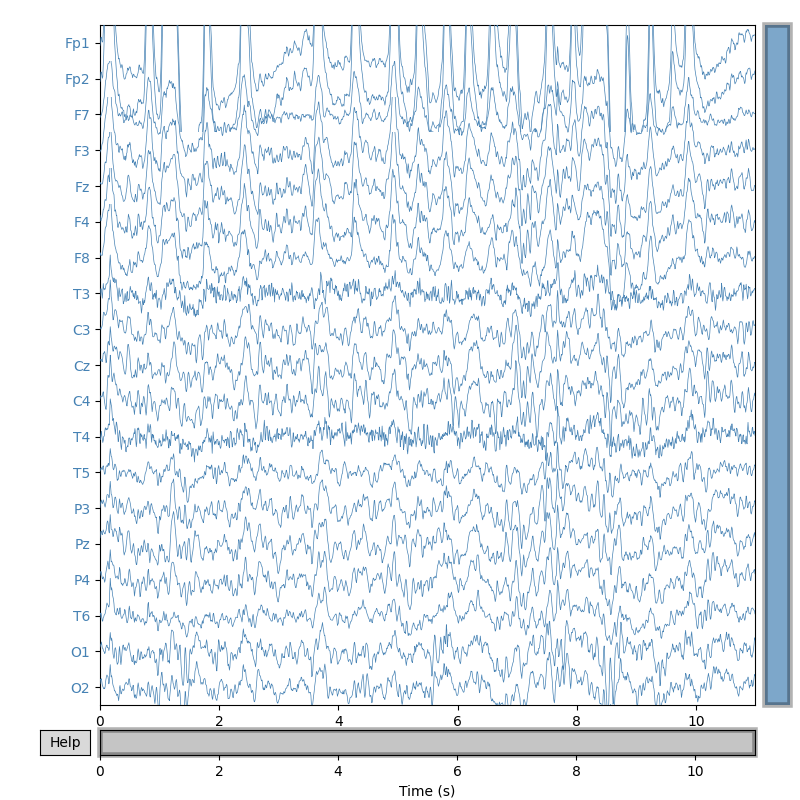
<MNEBrowseFigure size 800x800 with 4 Axes>
AJDC: Second-Order Statistics (SOS)-based BSS, diagonalizing cospectra¶
# Compute and diagonalize Fourier cospectral matrices between 1 and 32 Hz
window, overlap = sfreq, 0.5
fmin, fmax = 1, 32
ajdc = AJDC(window=window, overlap=overlap, fmin=fmin, fmax=fmax, fs=sfreq,
dim_red={'max_cond': 100})
ajdc.fit(signal_raw[np.newaxis, np.newaxis, ...])
freqs = ajdc.freqs_
# Plot cospectra in channel space, after trace-normalization by frequency: each
# cospectrum, associated to a frequency, is a covariance matrix
plot_cospectra(ajdc._cosp_channels, freqs, ylabels=ch_names,
title='Cospectra, in channel space')
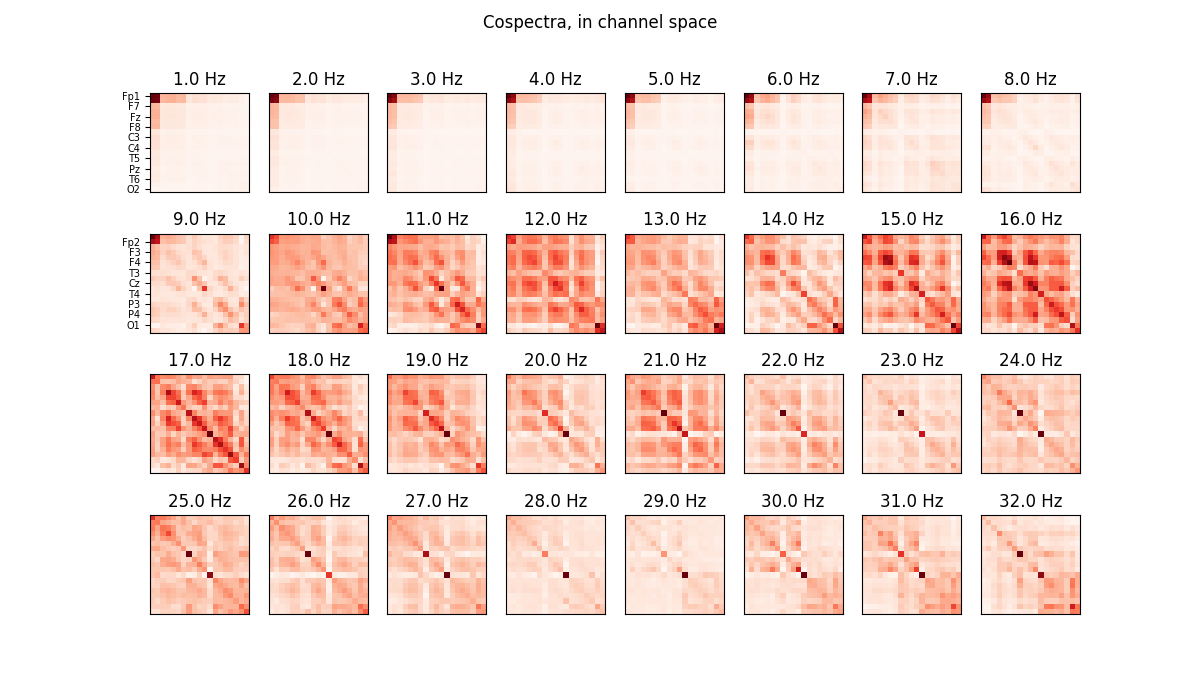
Condition numbers:
array([ 1. , 2.29766117, 4.09457756, 4.86981696,
6.09760458, 9.15865458, 13.21748535, 17.74436118,
26.1024296 , 27.31744246, 33.78134725, 45.22515539,
50.61007053, 60.36895283, 73.48533473, 74.73247287,
92.15600097, 121.30282659, 164.52162547])
Dimension reduction of Whitening on 17 components
<Figure size 1200x700 with 32 Axes>
# Plot diagonalized cospectra in source space
sr_count = ajdc.n_sources_
sr_names = ['S' + str(s).zfill(2) for s in range(sr_count)]
plot_cospectra(ajdc._cosp_sources, freqs, ylabels=sr_names,
title='Diagonalized cospectra, in source space')
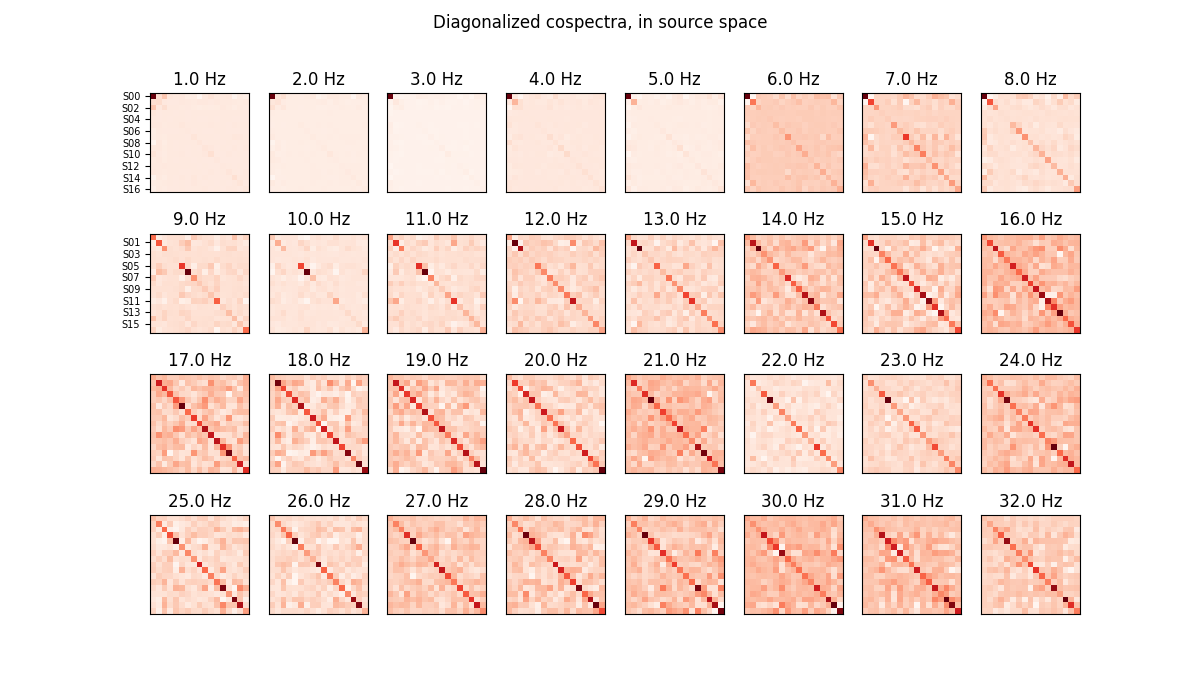
<Figure size 1200x700 with 32 Axes>
Source space¶
# Estimate sources S applying forward filters B to signal X: S = B X
source_raw = ajdc.transform(signal_raw[np.newaxis, ...])[0]
# Plot sources S
sr_info = create_info(ch_names=sr_names, ch_types=['misc'] * sr_count,
sfreq=sfreq)
source = RawArray(source_raw, sr_info, verbose=False)
source.plot(duration=duration, start=0, n_channels=sr_count,
scalings={'misc': 2e2}, title='EEG sources estimated by AJDC',
show_scalebars=False)
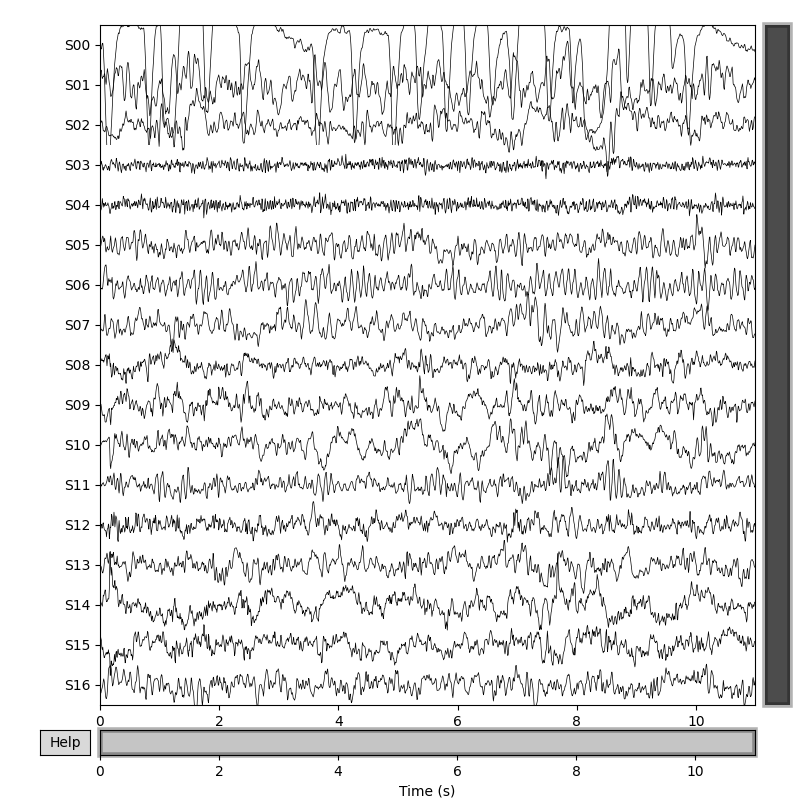
<MNEBrowseFigure size 800x800 with 4 Axes>
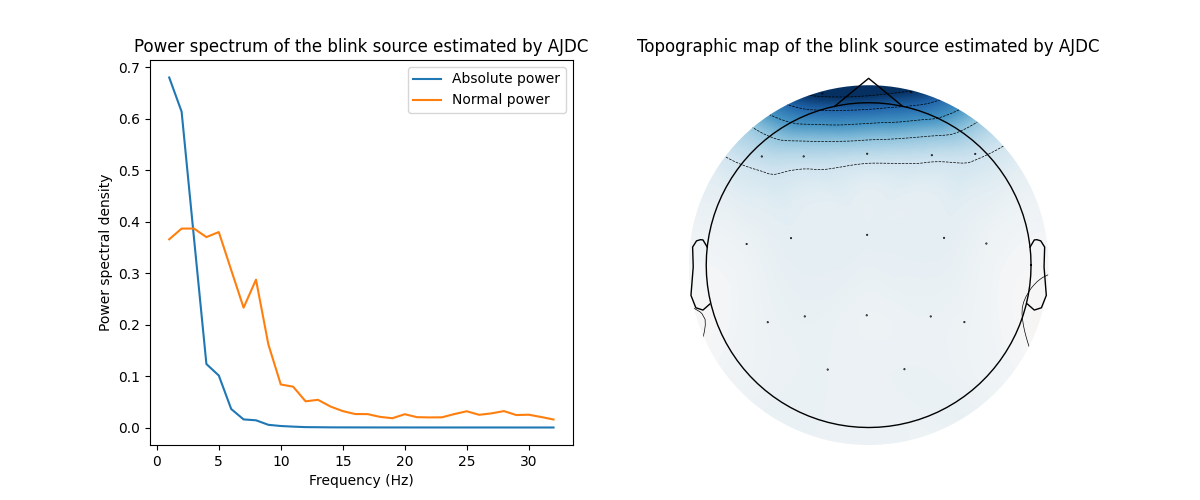
Artifact identification¶
# Identify artifact by eye: blinks are well separated in source S0
blink_idx = 0
# Get normal spectrum, ie power spectrum after trace-normalization
blink_spectrum_norm = ajdc._cosp_sources[:, blink_idx, blink_idx]
blink_spectrum_norm /= np.linalg.norm(blink_spectrum_norm)
# Get absolute spectrum, ie raw power spectrum of the source
f, spectrum = welch(source.get_data(picks=[blink_idx]), fs=sfreq,
nperseg=window, noverlap=int(window * overlap))
blink_spectrum_abs = spectrum[0, (f >= fmin) & (f <= fmax)]
blink_spectrum_abs /= np.linalg.norm(blink_spectrum_abs)
# Get topographic map
blink_filter = ajdc.backward_filters_[:, blink_idx]
# Plot spectrum and topographic map of the blink source separated by AJDC
fig, axs = plt.subplots(nrows=1, ncols=2, figsize=(12, 5))
axs[0].set(title='Power spectrum of the blink source estimated by AJDC',
xlabel='Frequency (Hz)', ylabel='Power spectral density')
axs[0].plot(freqs, blink_spectrum_abs, label='Absolute power')
axs[0].plot(freqs, blink_spectrum_norm, label='Normal power')
axs[0].legend()
axs[1].set_title('Topographic map of the blink source estimated by AJDC')
plot_topomap(blink_filter, pos=ch_info, axes=axs[1], extrapolate='box')
plt.show()
Artifact correction by BSS denoising¶
# BSS denoising: blink source is suppressed in source space using activation
# matrix D, and then applying backward filters A to come back to channel space
# Denoised signal: Xd = A D S
signal_denois_raw = ajdc.inverse_transform(source_raw[np.newaxis, ...],
supp=[blink_idx])[0]
# Plot denoised signal Xd
signal_denois = RawArray(signal_denois_raw, ch_info, verbose=False)
signal_denois.plot(duration=duration, start=0, n_channels=ch_count,
scalings={'eeg': 3e1}, color={'eeg': 'steelblue'},
title='Denoised EEG signal by AJDC', show_scalebars=False)
<MNEBrowseFigure size 800x800 with 4 Axes>
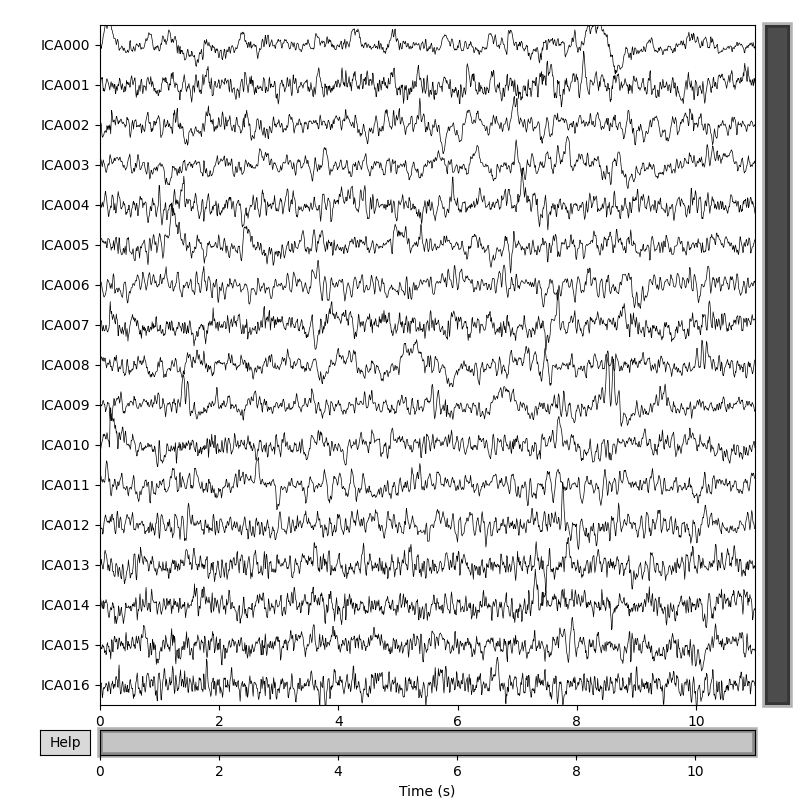
Comparison with Independent Component Analysis (ICA)¶
# Infomax-based ICA is a Higher-Order Statistics (HOS)-based BSS, minimizing
# mutual information
ica = ICA(n_components=ajdc.n_sources_, method='infomax', random_state=42)
ica.fit(signal, picks='eeg')
# Plot sources separated by ICA
ica.plot_sources(signal, title='EEG sources estimated by ICA')
# Can you find the blink source?
/home/docs/checkouts/readthedocs.org/user_builds/pyriemann/checkouts/latest/examples/artifacts/plot_correct_ajdc_EEG.py:161: RuntimeWarning: The data has not been high-pass filtered. For good ICA performance, it should be high-pass filtered (e.g., with a 1.0 Hz lower bound) before fitting ICA.
ica.fit(signal, picks='eeg')
Fitting ICA to data using 19 channels (please be patient, this may take a while)
Selecting by number: 17 components
Fitting ICA took 0.2s.
Creating RawArray with float64 data, n_channels=17, n_times=1408
Range : 0 ... 1407 = 0.000 ... 10.992 secs
Ready.
<MNEBrowseFigure size 800x800 with 4 Axes>
# Plot topographic maps of sources separated by ICA
ica.plot_components(title='Topographic maps of EEG sources estimated by ICA')
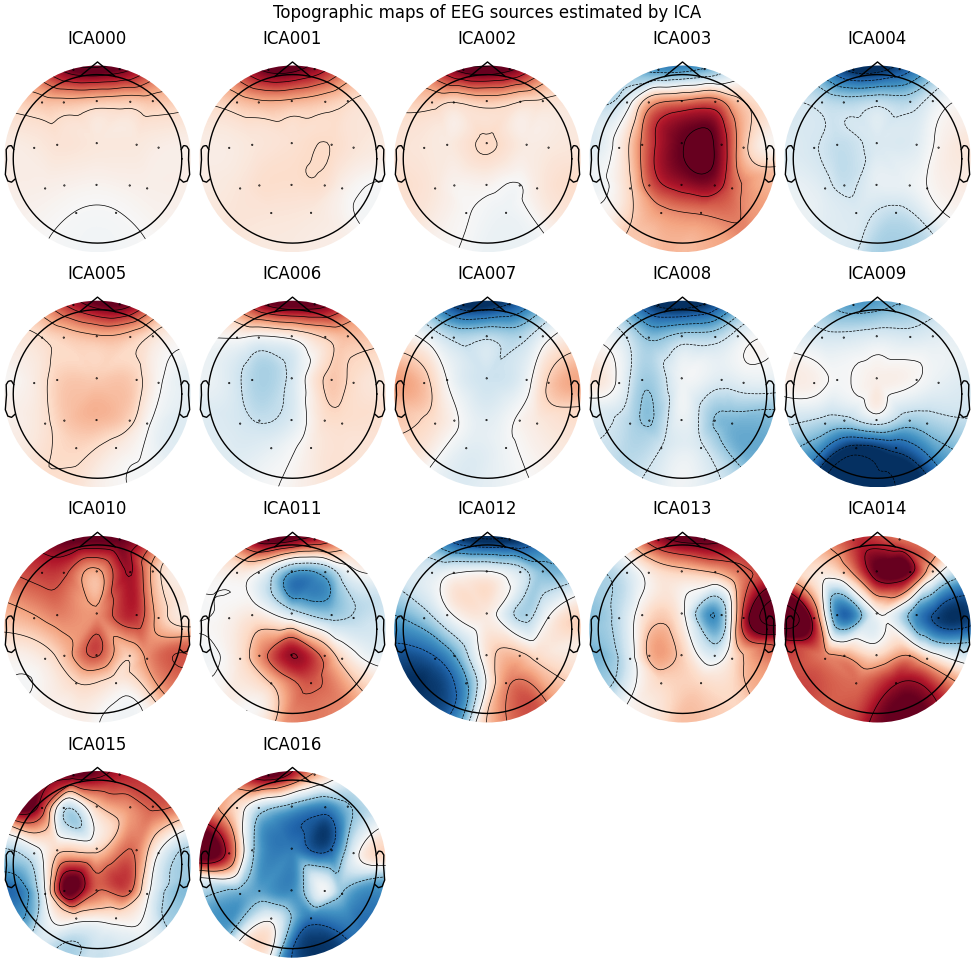
<MNEFigure size 975x967 with 17 Axes>
References¶
Total running time of the script: (0 minutes 10.040 seconds)