Note
Go to the end to download the full example code
Classifier comparison¶
A comparison of several classifiers on low-dimensional synthetic datasets, adapted to SPD matrices from [1]. The point of this example is to illustrate the nature of decision boundaries of different classifiers, used with different metrics [2]. This should be taken with a grain of salt, as the intuition conveyed by these examples does not necessarily carry over to real datasets.
The 3D plots show training matrices in solid colors and testing matrices semi-transparent. The lower right shows the classification accuracy on the test set.
# Authors: Quentin Barthélemy
#
# License: BSD (3-clause)
from functools import partial
from time import time
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
from sklearn.model_selection import train_test_split
from pyriemann.datasets import make_matrices, make_gaussian_blobs
from pyriemann.classification import (
MDM,
KNearestNeighbor,
SVC,
MeanField,
)
@partial(np.vectorize, excluded=['clf'])
def get_proba(cov_00, cov_01, cov_11, clf):
cov = np.array([[cov_00, cov_01], [cov_01, cov_11]])
with np.testing.suppress_warnings() as sup:
sup.filter(RuntimeWarning)
return clf.predict_proba(cov[np.newaxis, ...])[0, 1]
def plot_classifiers(metric):
figure = plt.figure(figsize=(12, 10))
figure.suptitle(f"Compare classifiers with metric='{metric}'", fontsize=16)
i = 1
# iterate over datasets
for ds_cnt, (X, y) in enumerate(datasets):
# split dataset into training and test part
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.4, random_state=42
)
x_min, x_max = X[:, 0, 0].min(), X[:, 0, 0].max()
y_min, y_max = X[:, 0, 1].min(), X[:, 0, 1].max()
z_min, z_max = X[:, 1, 1].min(), X[:, 1, 1].max()
# just plot the dataset first
ax = plt.subplot(n_datasets, n_classifs + 1, i, projection='3d')
if ds_cnt == 0:
ax.set_title("Input data")
# Plot the training points
ax.scatter(
X_train[:, 0, 0],
X_train[:, 0, 1],
X_train[:, 1, 1],
c=y_train,
cmap=cm_bright,
edgecolors="k"
)
# Plot the testing points
ax.scatter(
X_test[:, 0, 0],
X_test[:, 0, 1],
X_test[:, 1, 1],
c=y_test,
cmap=cm_bright,
alpha=0.6,
edgecolors="k"
)
ax.set_xlim(x_min, x_max)
ax.set_ylim(y_min, y_max)
ax.set_zlim(z_min, z_max)
ax.set_xticklabels(())
ax.set_yticklabels(())
ax.set_zticklabels(())
i += 1
rx = np.arange(x_min, x_max, (x_max - x_min) / 50)
ry = np.arange(y_min, y_max, (y_max - y_min) / 50)
rz = np.arange(z_min, z_max, (z_max - z_min) / 50)
print(f"Dataset n°{ds_cnt+1}")
# iterate over classifiers
for name, clf in zip(names, classifiers):
ax = plt.subplot(n_datasets, n_classifs + 1, i, projection='3d')
clf.set_params(**{'metric': metric})
t0 = time()
clf.fit(X_train, y_train)
t1 = time() - t0
t0 = time()
score = clf.score(X_test, y_test)
t2 = time() - t0
print(
f" {name}:\n training time={t1:.5f}\n test time ={t2:.5f}"
)
# Plot the decision boundaries for horizontal 2D planes going
# through the mean value of the third coordinates
xx, yy = np.meshgrid(rx, ry)
zz = get_proba(xx, yy, X[:, 1, 1].mean()*np.ones_like(xx), clf=clf)
zz = np.ma.masked_where(~np.isfinite(zz), zz)
ax.contourf(xx, yy, zz, zdir='z', offset=z_min, cmap=cm, alpha=0.5)
xx, zz = np.meshgrid(rx, rz)
yy = get_proba(xx, X[:, 0, 1].mean()*np.ones_like(xx), zz, clf=clf)
yy = np.ma.masked_where(~np.isfinite(yy), yy)
ax.contourf(xx, yy, zz, zdir='y', offset=y_max, cmap=cm, alpha=0.5)
yy, zz = np.meshgrid(ry, rz)
xx = get_proba(X[:, 0, 0].mean()*np.ones_like(yy), yy, zz, clf=clf)
xx = np.ma.masked_where(~np.isfinite(xx), xx)
ax.contourf(xx, yy, zz, zdir='x', offset=x_min, cmap=cm, alpha=0.5)
# Plot the training points
ax.scatter(
X_train[:, 0, 0],
X_train[:, 0, 1],
X_train[:, 1, 1],
c=y_train,
cmap=cm_bright,
edgecolors="k"
)
# Plot the testing points
ax.scatter(
X_test[:, 0, 0],
X_test[:, 0, 1],
X_test[:, 1, 1],
c=y_test,
cmap=cm_bright,
edgecolors="k",
alpha=0.6
)
if ds_cnt == 0:
ax.set_title(name)
ax.text(
1.3 * x_max,
y_min,
z_min,
("%.2f" % score).lstrip("0"),
size=15,
horizontalalignment="right",
verticalalignment="bottom"
)
ax.set_xlim(x_min, x_max)
ax.set_ylim(y_min, y_max)
ax.set_zlim(z_min, z_max)
ax.set_xticks(())
ax.set_yticks(())
ax.set_zticks(())
i += 1
plt.show()
Classifiers and Datasets¶
names = [
"MDM",
"k-NN",
"SVC",
"MeanField",
]
classifiers = [
MDM(),
KNearestNeighbor(n_neighbors=3),
SVC(probability=True),
MeanField(power_list=[-1, 0, 1]),
]
n_classifs = len(classifiers)
rs = np.random.RandomState(2022)
n_matrices, n_channels = 50, 2
y = np.concatenate([np.zeros(n_matrices), np.ones(n_matrices)])
datasets = [
(
np.concatenate([
make_matrices(
n_matrices, n_channels, "spd", rs, evals_low=10, evals_high=14
),
make_matrices(
n_matrices, n_channels, "spd", rs, evals_low=13, evals_high=17
)
]),
y
),
(
np.concatenate([
make_matrices(
n_matrices, n_channels, "spd", rs, evals_low=10, evals_high=14
),
make_matrices(
n_matrices, n_channels, "spd", rs, evals_low=11, evals_high=15
)
]),
y
),
make_gaussian_blobs(
2*n_matrices, n_channels, random_state=rs, class_sep=1., class_disp=.5,
n_jobs=4
),
make_gaussian_blobs(
2*n_matrices, n_channels, random_state=rs, class_sep=.5, class_disp=.5,
n_jobs=4
)
]
n_datasets = len(datasets)
cm = plt.cm.RdBu
cm_bright = ListedColormap(["#FF0000", "#0000FF"])
Classifiers with Riemannian metric¶
plot_classifiers("riemann")
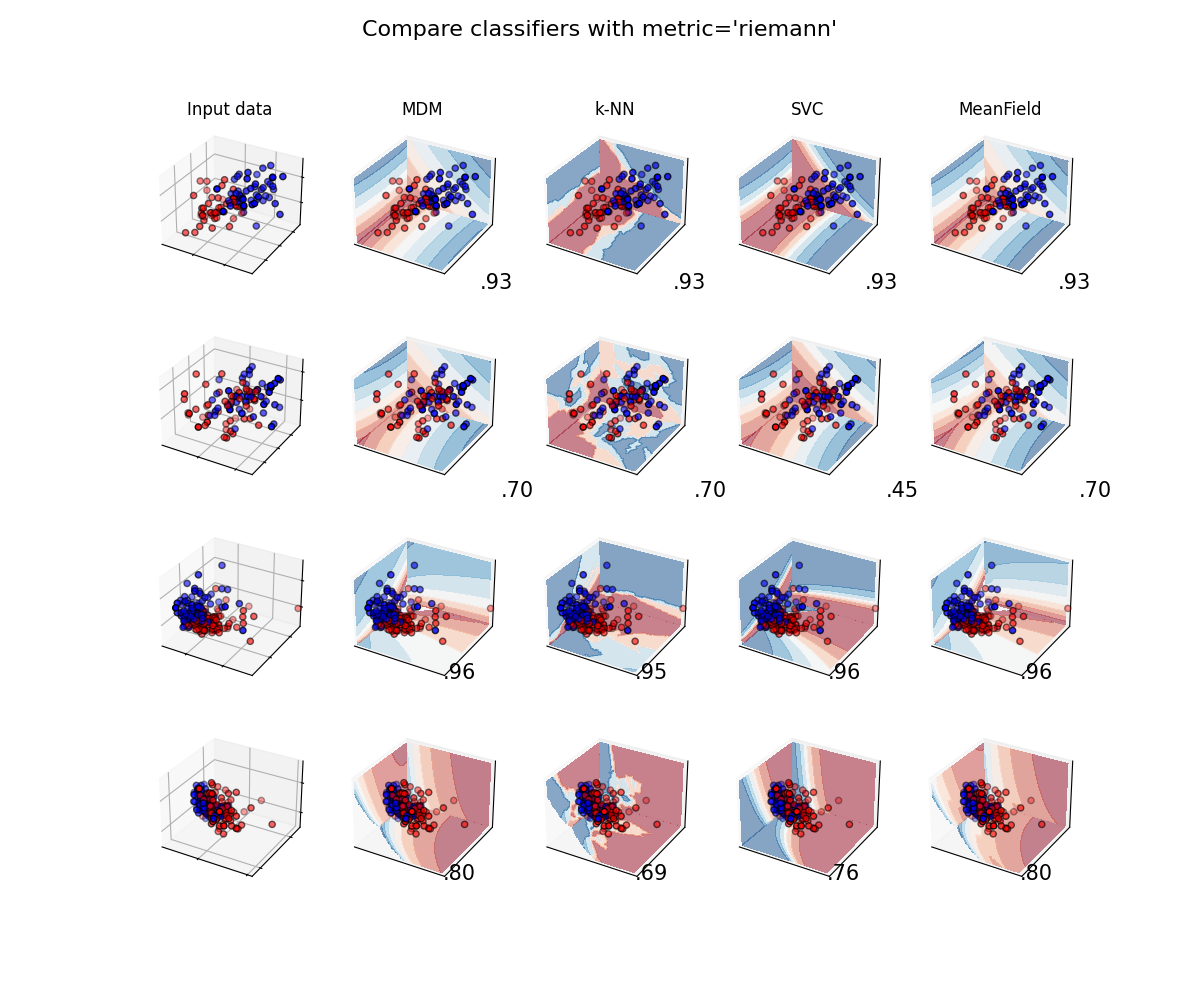
Dataset n°1
MDM:
training time=0.00293
test time =0.00681
k-NN:
training time=0.00006
test time =0.13814
SVC:
training time=0.00460
test time =0.00197
MeanField:
training time=0.00338
test time =0.02028
Dataset n°2
MDM:
training time=0.00281
test time =0.00662
k-NN:
training time=0.00006
test time =0.13819
SVC:
training time=0.00446
test time =0.00192
MeanField:
training time=0.00619
test time =0.01830
Dataset n°3
MDM:
training time=0.00596
test time =0.01214
k-NN:
training time=0.00006
test time =0.39111
SVC:
training time=0.00737
test time =0.00211
MeanField:
training time=0.00661
test time =0.03868
Dataset n°4
MDM:
training time=0.00590
test time =0.01205
k-NN:
training time=0.00006
test time =0.39407
SVC:
training time=0.00764
test time =0.00225
MeanField:
training time=0.00657
test time =0.03866
Classifiers with Log-Euclidean metric¶
plot_classifiers("logeuclid")
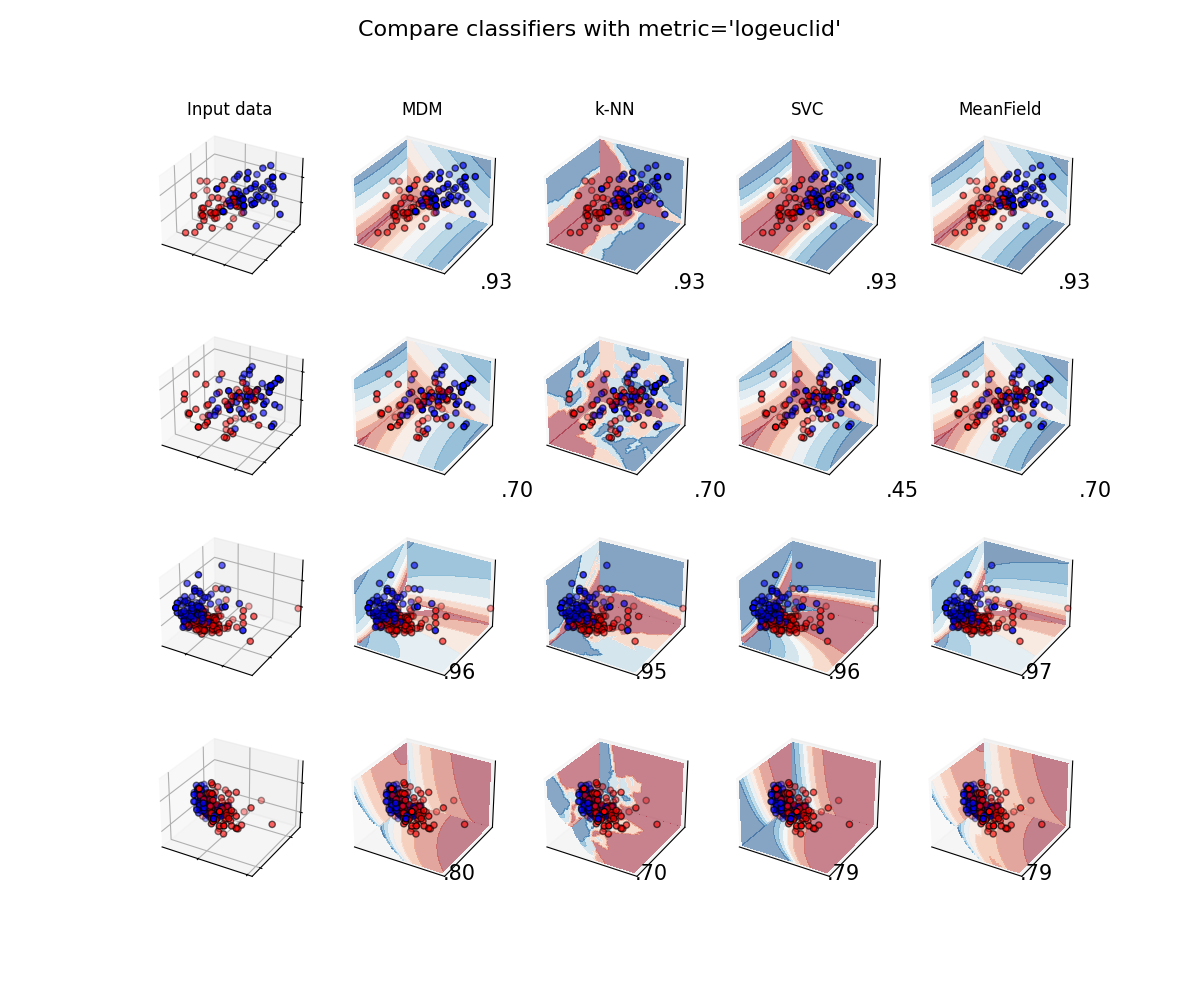
Dataset n°1
MDM:
training time=0.00084
test time =0.00980
k-NN:
training time=0.00006
test time =0.18602
SVC:
training time=0.00279
test time =0.00174
MeanField:
training time=0.00344
test time =0.03058
Dataset n°2
MDM:
training time=0.00086
test time =0.00975
k-NN:
training time=0.00006
test time =0.18779
SVC:
training time=0.00283
test time =0.00174
MeanField:
training time=0.00343
test time =0.03062
Dataset n°3
MDM:
training time=0.00094
test time =0.01847
k-NN:
training time=0.00006
test time =0.59480
SVC:
training time=0.00333
test time =0.00190
MeanField:
training time=0.00667
test time =0.05992
Dataset n°4
MDM:
training time=0.00098
test time =0.01835
k-NN:
training time=0.00005
test time =0.63659
SVC:
training time=0.00416
test time =0.00193
MeanField:
training time=0.00657
test time =0.05915
Classifiers with Euclidean metric¶
plot_classifiers("euclid")
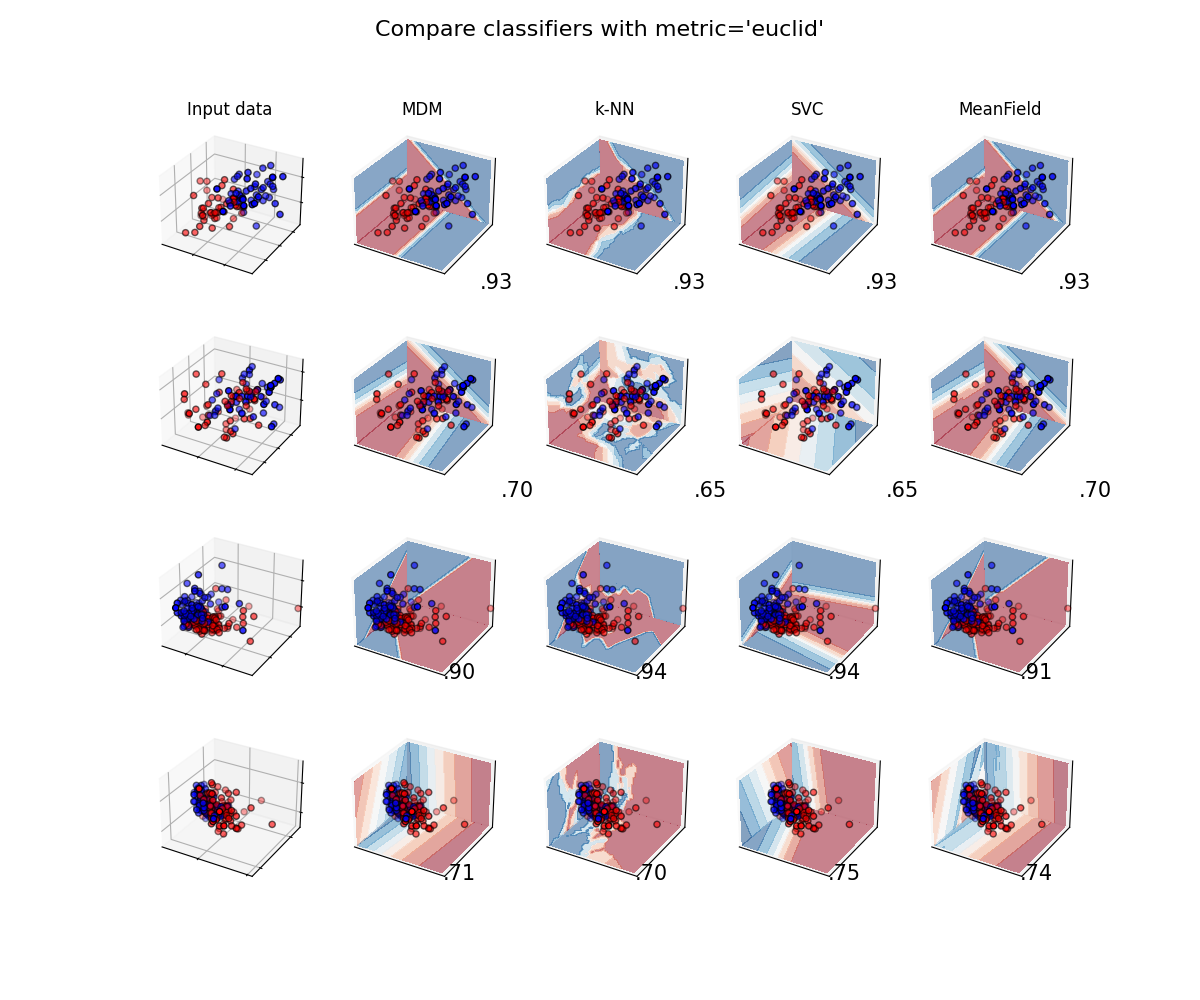
Dataset n°1
MDM:
training time=0.00039
test time =0.00249
k-NN:
training time=0.00006
test time =0.04487
SVC:
training time=0.00191
test time =0.00137
MeanField:
training time=0.00343
test time =0.00774
Dataset n°2
MDM:
training time=0.00040
test time =0.00249
k-NN:
training time=0.00006
test time =0.04506
SVC:
training time=0.00262
test time =0.00132
MeanField:
training time=0.00342
test time =0.00767
Dataset n°3
MDM:
training time=0.00043
test time =0.00381
k-NN:
training time=0.00006
test time =0.13714
SVC:
training time=0.00356
test time =0.00137
MeanField:
training time=0.00654
test time =0.01371
Dataset n°4
MDM:
training time=0.00040
test time =0.00383
k-NN:
training time=0.00006
test time =0.13670
SVC:
training time=0.00321
test time =0.00141
MeanField:
training time=0.00675
test time =0.01398
References¶
Total running time of the script: (6 minutes 38.003 seconds)